6. Phylogenetics and
taxonomy of Santalales
|

 |
The first molecular phylogeny of the sandalwood order
(Santalales) was published over 20 years ago (Nickrent and
Franchina 1990) and since those humble beginnings my
lab and collaborators have generated much information about
the evolution of this fascinating order of plants. Some
of the questions that our group has addressed include 1) how
many times has parasitism evolved? 2) how many times has the
mistletoe habit evolved? 3) are the polymorphic families
Santalaceae and Olacaceae monophyletic? and 4) what are the
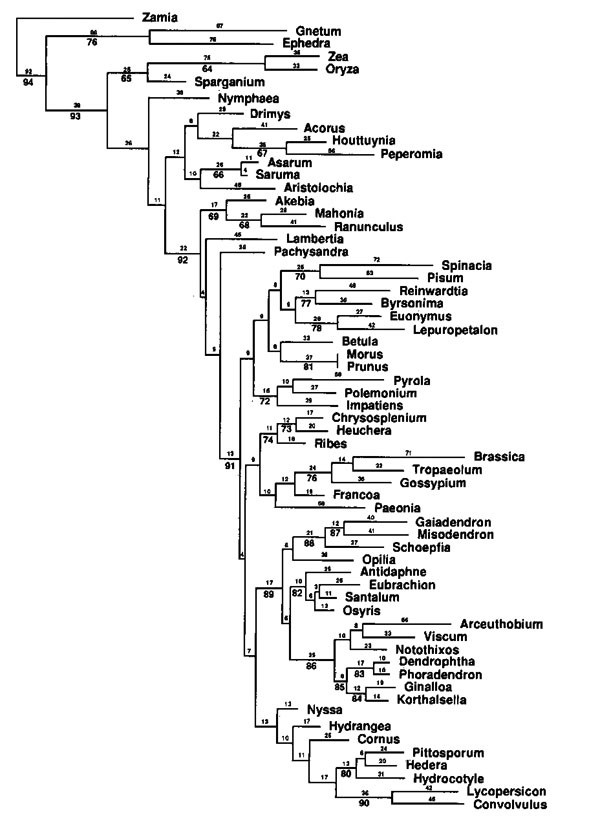
relationships among the genera in the large family
Loranthaceae?
Every group traditionally recognized as a family has been
examined using molecular techniques. This work was
possible only through the collaborative efforts of these
individuals: Olaceae (Dr. Valéry Malécot), Loranthaceae
(Drs. Romina Vidal-Russell and Guillermo Amico),
Misodendraceae (Dr. Vidal-Russell) and Santalaceae (Drs.
Joshua Der and Miguel A. García). The Taxon
publication (Nickrent et al. 2010) provided a complete
synopsis of current understanding of relationships in the
order, and gave a revised classification based mainly on the
molecular evidence.
During the course of this work, four new genera were named (Hondurodendron,
Lacomucinaea, Pilgerina,
and Staufferia).
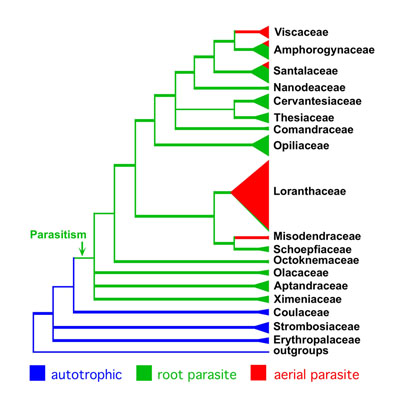
The reclassification of the order based on the concept
of monophyly resulted in some new (or recycled) family
names. Olacaceae was split into Aptandraceae,
Coulaceae, Erythropalaceae, Octoknemaceae, Olacaceae s.
s., Schoepfiaceae, Strombosiaceae, and Ximeniaceae.
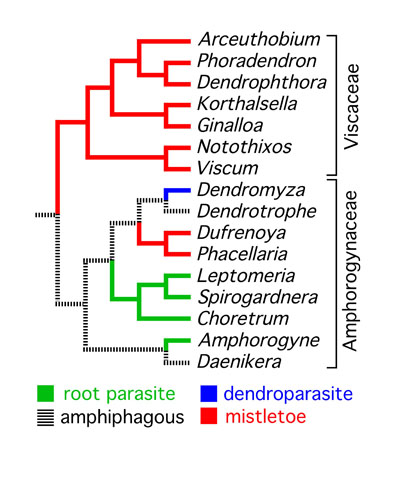
Santalaceae was split into Amphorogynaceae,
Cervantesiaceae, Comandraceae, Nanodeaceae, Santalaceae
s. st., and Thesiaceae. Three genera
formerly classified in Santalaceae were moved to
Schoepfiaceae (Arjona,
Quinchamalium) and
Opiliaceae (Anthobolus).
Excluding
Balanophoraceae, the order now consists of 18 families, 148
genera and nearly 2300 species. Our classification of
Olacaceae was adopted by the Angiosperm Phylogeny Group (APG
III 2009) but unfortunately they did not follow our
recommendations for "Santalaceae s. lat." and chose to lump
Viscaceae into it. My viewpoint on this was expressed in the
Haustorium newsletter found on pp. 4-6 HERE.
Answers to some of the questions raised above can now be
given. It appears that parasitism evolved just once in the
order, although more recent multigene analyses draw this
conclusion into question (stay tuned!). It seems
clear, however, that the mistletoe habit evolved five times
independently. "Santalaceae" and "Olacaceae", as
traditionally defined, were polyphyletic. In our new
classification all families are monophyletic. Thanks
to the efforts of Romina Vidal-Russell, we now have a
phylogeny for Loranthaceae. These results allowed us
to proposed the first infrafamilial classification of the
family based on phylogenetic evidence. Nuytsia
is sister to all other genera in the family and base
chromosome number shows a progressive aneuploid reduction
from X=12 in basal genera to X=9 in more derived clades
(e.g. the African members). These data literally
turned a previous biogeographic concept on its head,
highlighting dispersal, not vicariance, for the origin of
the African and Asian loranths. |
Publications:
- Nickrent, D. L., F. Anderson,
& J. Kuijt. 2019. Inflorescence evolution in
Santalales: Integrating morphological characters and
molecular phylogenetics. American Journal of Botany
106(3):402-414. For a
pdf file of this article click HERE.
- Nickrent D. L. 2017. Status of
the genera Colpoon, Osyris and Rhoiacarpos
in South Africa. Bothalia: African Biodiversity &
Conservation 47(1): (online 13 Nov. 2017). For
a PDF file of this article click
HERE.
- Nickrent, D. L. 2016.
Ximeniaceae, Schoepfiaceae, Comandraceae, Thesiaceae,
Cervantesiaceae, Santalaceae, Viscaceae. Pp.
404-440 in: Flora North America, Volume 12, Flora
North America Editorial Committee (eds.), Oxford
University Press, New York. For
a
PDF file of this article click HERE.
These treatments are available online HERE.
- Devkota, M. P., J. Macklin, &, D.
L. Nickrent. 2015. The status of the
mistletoe genus Dufrenoya
Chatin (Amphorogynaceae) with a specific focus on
Nepal. Flora 215:75-83. For
a PDF file of this article click
HERE.
- Su H.-J., J.-M. Hu, F. E. Anderson and D.
L. Nickrent. 2015. Phylogenetic
relationships of Santalales with insights into the
origins of holoparasitic Balanophoraceae. Taxon 64(3):
491-506. For a
PDF file of this article click
HERE.
- Nickrent,
D. L.
2011. Santalales (Including Mistletoes).
Encyclopedia of Life Science. John Wiley & Sons,
Ltd.: Chichester [DOI:
10.1002/9780470015902.a0003714.pub2]. Wiley website
(search for article) HERE.
For a PDF file of this
article click HERE.
- Ulloa, C. U, D.
L. Nickrent , C. Whitefoord, and D. L. Kelly.
Hondurodendron,
a new monotypic genus of Aptandraceae from Honduras.
Annals of the Missouri Botanical Garden 97: 457-467.
For a PDF file of this article click HERE.
- Nickrent, D. L.
V. Malécot, R. Vidal-Russell, and J. P. Der. 2010. A
revised classification of Santalales. Taxon 59:
538-558. For
a PDF file of this article click HERE.
Supplemental data file on chromosome
numbers HERE.
- Vidal-Russell, R. and D.
L.
Nickrent. 2008. Evolutionary relationships in
the showy mistletoe family (Loranthaceae). American
Journal of Botany 95: 1015-1029. For a PDF
file of this article click HERE.
- Rogers, Z. S., D.
L. Nickrent, and V. Malécot. 2008. Staufferia
and Pilgerina: two new arborescent genera of
Santalaceae from Madagascar. Annals of the Missouri
Botanical Garden 95: 391-404. For a PDF file of
this article click HERE.
- Vidal-Russell, R. and D.
L. Nickrent. 2008. The first mistletoes:
origins of aerial parasitism in Santalales. Molecular
Phylogenetics and Evolution 47 (2): 523-537. For a PDF
file of this article (constructed from original
files, not MPE pdf that is restricted by Elsevier),
click HERE.
- Der, J. P. and Nickrent,
D. L. 2008. A molecular phylogeny of
Santalaceae (Santalales). Systematic Botany 33:
107-116. For a PDF file of this article, click HERE.
- Malécot, V. and Nickrent,
D. L. 2008. Molecular phylogenetic
relationships of Olacaceae and related Santalales.
Systematic Botany 33: 97-106. For a PDF file of
this article, click HERE.
- Malécot, V., D.
L. Nickrent, P. Baas, L. van den Oever, D.
Lobreau-Callen. 2004. Phylogeny of Olacaceae based on
a morphological cladistic analysis. Systematic Botany.
29:569-586. For a PDF file of this article, click HERE
- Nickrent, D. L.,
and V. Malécot. 2001. A molecular phylogeny of
Santalales. Pages 69-74 in A. Fer, P. Thalouarn, D. M.
Joel, L. J. Musselman, C. Parker, and J. A. C.
Verkleij, eds. Proceedings of the 7th. International
Parasitic Weed Symposium. Faculté des Sciences,
Université de Nantes, Nantes, France. For a PDF file
of this article, click HERE.
An updated web version of this work is HERE.
- Nickrent, D. L. and R. J. Duff. 1996. Molecular
studies of parasitic plants using ribosomal RNA. Pp.
28-52. In: M. T. Moreno, J. I. Cubero, D. Berner, D.
Joel, L. J. Musselman, C. Parker (eds.), Advances in
Parasitic Plant Research, Junta de Andalucia,
Dirección General de Investigación Agraria, Cordoba,
Spain. For PDF file of this article, click HERE.
- Nickrent, D. L.
and C. R. Franchina. 1990. Phylogenetic relationships
of the Santalales and relatives. Journal of Molecular
Evolution 31: 294-301. For a PDF file of the article,
click HERE.
|
|